
| MIPLCC Documentation |
| version 0.0 |
| Table of Contents |
 |
|
|
| The MIPL Core Code (MIPL-CC) is a collection of C++ and bash programs for the simulation, optimization, processing, and analysis of multimodal and multiparametric neuroimaging data. The entire MIPL-CC is made available to the general public via the IU Software License. Basically, you can use MIPL-CC for free for non-commercial research. If you want to use it to conduct for-profit research, or to build products you will sell, you have to negotiate with Indiana University. No warranties of any kind are given, and you use the code at your own risk. |
This manual organizes the various programs of the MIPL-CC by functional sections. We respectfully ask that you cite the appropriate references any time you use MIPL-CC. The correct references are given in each section. |
You can download the MIPL-CC source code and binaries from the data & code repository. |
Special care is taken to ensure the MIPL-CC works in harmony with FSL, and you should be able to run both in the same environment (and on the same data) without issue. |
All of our programs print a help style output when you call them without an argument - this is the best way to quick information on the correct way to run our programs. |
|
|
| There are 2 ways to install MIPLCC: 1) the binary way, and 2) the compile way. The compile way is really only necassary if you're trying to install in an environment that doesn't already run FSL, or if you feel the need to attempt to optimize the binaries in a way that is specific to your architecture. |
The binary way. Download the MIPLCC binaries from the data & code folder - they are all contained in the file MIPLCC_binaries.tar.gz. Extract this file into your FSL environment in /usr/bin/ or $HOME/bin/. Extract the file with the following command: |
tar -xzf MIPLCC_binaries.tar.gz |
This will create a new directory in /user/bin/ or $HOME/bin/ called MIPLCC_binaries. That's it - you can now us the MIPLCC by calling the individual programs from the command line. |
The compile way. Download the MIPLCC source code from the data & code folder - they are all contained in the file MIPLCC_source.tar.gz. Extract this file into a working directory of your choice with the following command: |
tar -xzf MIPLCC_source.tar.gz |
This will create a new directory called MIPLCC_source. You will now need to compile each program you wish to use individually - there are no makefiles but you will find an example compile command at the top of each .cpp file. Generally you can compile with the g++, Cray C++, and/or Intel C++ compilers without issue as long as you include the C++17 standard (i.e. -std=c++17) and the proper flags for zlib, MPICH, and the General_MIPL library. Once compiled, copy the resulting binaries to /user/bin/ or $HOME/bin/. |
|
|
|
|
|
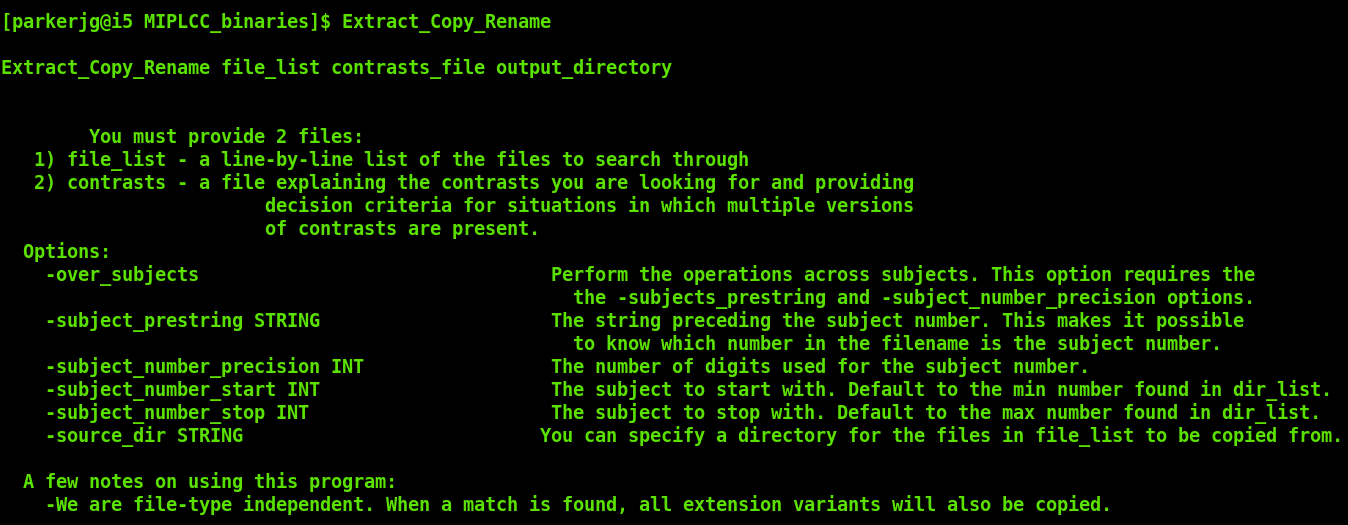
Extract_Copy_Rename is used mainly at the beginning of a retrospective study to search through a list of acquisitions to identify, copy, and rename the specific modalitiies/sequences/parametric maps you need for the experiment. To run Extract_Copy_Rename you must provide 2 files: |
|
1) file_list - a line-by-line list of the files to search through, and 2) contrasts - a file explaining the contrasts you are looking for and providing decision criteria for situations in which multiple versions of contrasts are present. |
|
Normally you generate file_list by listing all of the files in a target directory and redirecting the output to a text file: |
ls all_my_nifti_data/ > file_list.txt |
|
Here is an example file_list. The contrasts file lists each of the sequences and their descriptors/keywords to look for. Under each name, an ordered list provides the keywords that should be searched for. As soon as an entire keyword line is matched, the search for that sequence ends. Thus, the order of the lines indicates goes from more desirable (higher up) to less desirable (lower down) sequences. Specify the beginning of a sequence by the Name: operator. For example, Name: T1w would be a good name for the T1 anatomical. The following lines then indicate each combination of keywords that should be included (+) or excluded (-) When the Name: operator is given the string GLOBAL, the following tags will be applied to all entries. Comments are indicated by #. Here is an example example_contrasts_file. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|